Research

Moran, Graham
Title/s: Professor
Carl Moore Endowed Research Chair
Specialty Area: Biochemistry
Office #: FH-419
Phone: 773.508.3756
Email: gmoran3@luc.edu
Research Interests
The Moran research group is interested in finding evidence for the chemical mechanisms of enzymes. Enzymes offer many inherently fascinating intricate puzzles that can be solved by systematic observation. As catalysts, they tend to carry out highly specific chemistry often adhering to a single reaction coordinate. We employ an array of methods to study enzyme reactions and tend to more often use transient state kinetics to observe the chemistry. Transient state or pre-steady state kinetics is a highly diagnostic and more explicit approach compared other some methods. Seeing chemistry happen on the surface of an enzyme in real time facilitates constructing a description of the transformation at work. We have generally applied such methods to systems that are amenable observed directly (due to changes in a chromophore or fluorophore with catalysis), however, many enzyme systems are regarded as being opaque to such methods due to a lack of tractable spectrophotometric signals. In recent years, we have studied a number of “opaque” enzymes in real time using kinetically coupled validated reporter methods. These methods have greatly expanded the number of systems we can study, allowing us to choose projects largely based on the chemistry catalyzed as opposed to how tractable the system is to study with our methods.
While we have studied many enzymes over the last two decades, our primary interests in the near to mid-future are the enzymes of riboflavin biosynthesis, the enzymology of cytolsolic isocitrate dehydrogenase and dihydropyrimidine dehydrogenase.
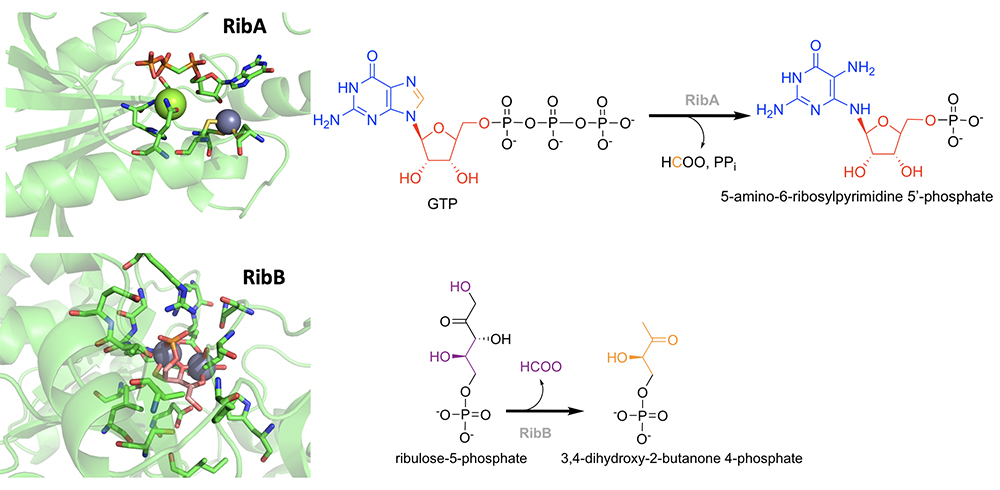
1. Riboflavin Biosynthesis. Riboflavin biosynthesis includes five enzymatic activities that are largely unparalleled in nature. All organisms require riboflavin, but many higher organisms do not have the enzymatic machinery to generate it. Riboflavin biosynthetic enzymes have been studied in sufficient detail to establish that these enzymes do unprecedented chemistry, but the chemical mechanisms offered for them have numerous unanswered questions. We intend initially to study the two entry points of the pathway through which GTP and ribulose-5-phosphate are inducted. RibA does an unusual deformylation that excises the bridging methyne (C8) of the imidazole heterocycle of the GTP guanosine while also hydrolyzing pyrophosphate. RibB is being studied collaboratively with the laboratory of Audrey Lamb at the University of Kansas. This enzyme catalyzes a ribulose-5-phosphate rearrangement that oddly eliminates the (internal) 4 carbon as formate. Therefore, we propose to investigate the underlying chemistry of these two mechanistically understudied deformylation reactions of riboflavin biosynthesis. We propose the development of innovative enzyme-specific, validated transient state kinetic approaches that are augmented with structural biology data. Each of the experimental approaches outlined below take advantage of the extraordinarily slow turnover numbers for both enzymes (0.002-0.05 s-1, or 7 to 180 turnovers per hour). The objective is to reveal previously uncharacterized steps in the catalytic cycle of RibA and RibB. These data will be obtained in parallel with specific, ligand-bound structures of the enzymes that are relevant to the mechanistic data obtained.
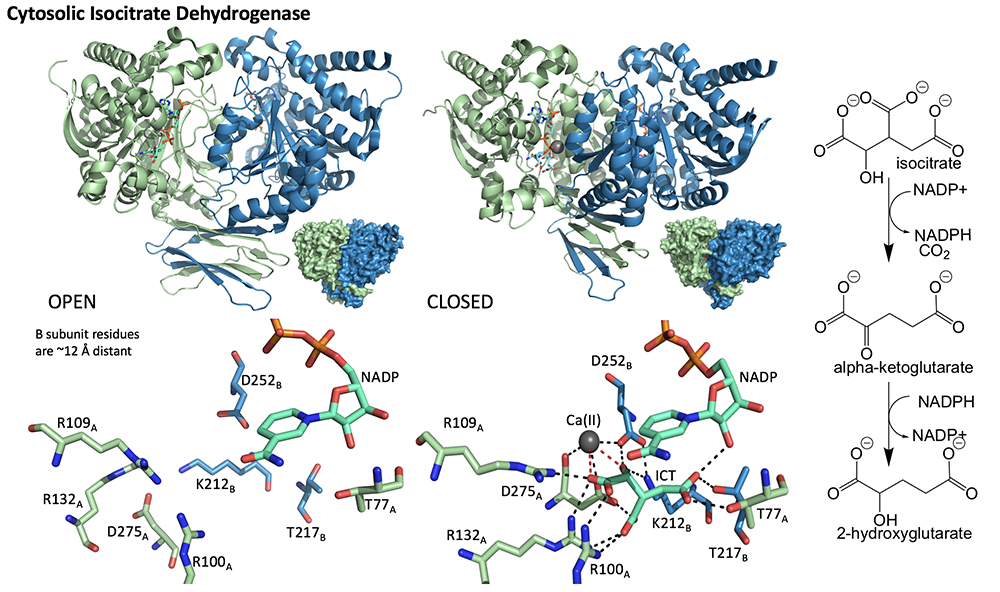
2. Cytosolic Isocitrate Dehydrogenase. Specific mutations in the cytosolic NADP-dependent isocitrate dehydrogenase (ICDH) result in a neomorphic activity that permits the enzyme to catalyze the reduction of the normal product of the reaction, alpha-ketoglutarate (AKG) to D-2-hydroxyglutarate (2HG). 2HG is an onco-metabolite that disrupts the activity of numerous AKG-dependent enzymes that groom access to the genome thereby inducing proliferative behavior. While D-2HG-dehydrogenase (2HGD) is a constitutive component of mammalian cells that would detoxify 2HG by restoring AKG, this enzyme for unknown reasons does not prevent accumulation of 2HG. We intend to study the interplay of ICDH with 2HGD. While ICDH has been studied extensively there remains a great deal to be unearthed in terms of ligand interactions. Moreover, 2HGD is entirely unknown in the literature and awaits complete detailed mechanism and structural characterization.
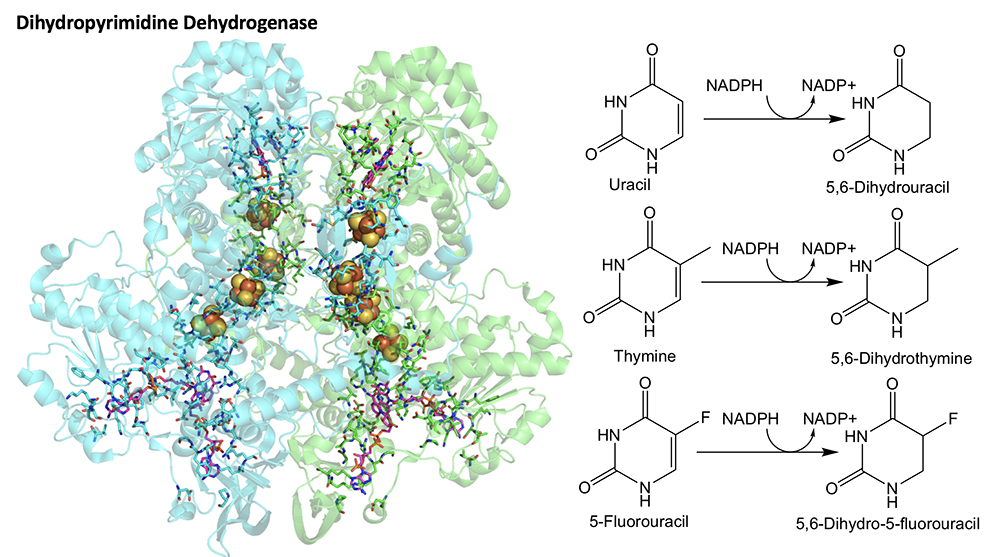
3. Dihydropyrimidine Dehydrogenase. Dihydropyrimidine dehydrogenase (DPD) is an enzyme involved in the degradation of pyrimidine bases. It reduces unsaturated bonds in the uracil and thymine heterocycles. It also has an important therapeutic function in the detoxification of 5-fluorouracil, a common chemotherapeutic agent. While the reaction catalyzed by DPD is simple and involves just the transfer of two electrons from NADPH to Uracil, the structure of DPD is rather complex. In addition to existing as a homodimer of ca. 1000 amino acid subunits, DPD also utilizes FAD, FMN and four Fe-S centers. Precisely how these redox cofactors are involved in the reaction is largely unknown as no direct evidence for the function of the Fe-S centers has been demonstrated despite the fact that they appear to bridge between the FAD and FMN cofactors. DPD has a significant number of spectrophotometric handles that should allow its mechanism and kinetics to be readily studied. We propose to strictly control molecular oxygen concentration to ensure that the chemistry observed is solely that of the native catalytic function.
The Moran Laboratory

Selected Publications
- Sebastian Flieger, Mao Takagaki, Natsuko Kondo, Marlon R. Lutz, Jr., Yash Gupta, Graham Moran, Prakasha Kempaiah, Narayan Hosmane, and Daniel P. Becker. (2023) Carborane-Containing Hydroxamate MMP Ligands for the Treatment of Cancers using Boron Neutron Capture Therapy (BNCT) Efficacy without Tumor Cell Entry. International Journal of Molecular Sciences 24, 10.3390/ijms24086973
https://www.mdpi.com/1422-0067/24/8/6973
- Sida Shen, Arseniy Butrin, Brett A. Beaupre, Glaucio M. Ferreira, Peter F. Doubleday, Daniel H. Grass, Wei Zhu, Neil L. Kelleher, Graham R. Moran, Dali Liu, and Richard B. Silverman. (2023) Structural and Mechanistic Basis for the Inactivation of Human Ornithine Aminotransferase by 3-Amino-4-fluorocyclopentenecarboxylic Acid. Molecules 28(3), 1133.
https://pubmed.ncbi.nlm.nih.gov/36770800/
- Madison M. Smith, Dariush C. Forouzesh, Nicholas E. Kaley, Dali Liu and Graham R. Moran. (2023)Mammalian Dihydropyrimidine Dehydrogenase: Answers to Outstanding Mechanistic Questions from Transient State Analysis of Charge Transfer Complexes. Archives of Biochemistry and Biophysics, 736, 109517.
https://pubmed.ncbi.nlm.nih.gov/36681231/
- Madison M. Smith and Graham R. Moran (2023) The Unusual Chemical Sequences of Dihydropyrimidine Dehydrogenase Revealed by Transient State Analysis. Methods in Enzymology. 685:373-403.
https://pubmed.ncbi.nlm.nih.gov/37245908/ - Madison M Smith, Tyler B. Alt, David L. Williams and Graham R. Moran (2023) Descriptive Analysis of Transient State Observations of Thioredoxin Glutathione Reductase (Sec597Cys) from Schistosoma mansoni. Biochemistry 62(9):1497-1508.
https://pubmed.ncbi.nlm.nih.gov/37071546/ - Madison M. Smith and Graham R. Moran (2023) Assigning Function to Active Site Residues of Schistosoma mansoni Thioredoxin/Glutathione Reductase from Analysis of Transient State Reductive Half-Reactions with Variant Forms of the Enzyme. Frontiers in Biomolecular Sciences. Volume 10
https://doi.org/10.3389/fmolb.2023.1258333 - Tyler B. Alt, Matthew Hoag and Graham R. Moran (2023) Bacterial Dihydropyrmidine Dehydrogenase: Transient State Analysis Reveals Both Reductive Activation Prior to Turnover and Diminished Substrate Effector Roles Relative to the Mammalian Form. Archives of Biochemistry and Biophysics 748:109772.
https://www.sciencedirect.com/science/article/pii/S0003986123002710 - Devitt, Allison; Vargas, Abigail; Zhu, Wei ; Des Soye, Ben; Butun, Fatma; Alt, Tyler; Kaley, Nicholas; Ferreira, Glaucio; Moran, Graham R.; Kelleher, Neil; Liu, Dali; Silverman, Richard. (2024) Design, Synthesis, and Mechanistic Studies of (R)-3-Amino-5,5-difluorocyclohex-1-ene-1-carboxylic Acid as an Inactivator of Human Ornithine Aminotransferase. ACS Chemical Biology. 19:1066-1081.
https://pubs.acs.org/doi/10.1021/acschembio.4c00022 - Madison M. Smith and Graham R. Moran (2024) Building on a theme: The Redox Hierarchy of Flavoprotein Disulphide Reductases. Archives of Biochemistry and Biophysics. 775:109966.
https://www.sciencedirect.com/science/article/pii/S0003986124000857 - Corine O. Smith and Graham R. Moran (2024) Elucidation of the Catalytic Sequence of Dihydroorotate Dehydrogenase B from Lactoccocus lactis: Evidence for Accumulation of a Flavin Bi-semiquinone State in Catalysis. Biochemistry 63:1347-1358
https://pubs.acs.org/doi/10.1021/acs.biochem.4c00025
